- Introduction
- The Data
- The Model
- Results and Interpretation
- Deployment
Introduction
print(fiction) is a solo project I worked on to explore the data on and around fictional stories.
I used Scrapy to scrape metadata for over 20,000 books from GoodReads and used it to train a series of machine learning classifiers. The final version of the model classified books as either fiction or nonfiction with ~80% accuracy.
The dataset is freely available for download on GitHub.
I built an interactive dashboard using Plotly Dash that can be used to tinker with the model parameters and view the resulting prediction in real time.
This project is part of an on-going series of exploratory articles and projects called Sci-Fi IRL, through which I am exploring the relationship between science-fiction and the real world. It is my belief that the stories we read, write, and believe in, particularly about our future, have an effect on how that future ultimately turns out.
Our human minds are geared toward thinking about what could go wrong. It follows that the majority of stories in popular and niche culture are written about how things could go wrong, usually catastrophically so—it "tells a good story".
In the case of science-fiction, stories tend to be dystopian in nature, showing what could go wrong if our technology advances along certain trajectories.
But does this affect our outlook on what these technologies can do for us?
While it is always good to consider the possible ramifications of technological advances, I believe that too many dystopian stories are causing humans, as a civilization, to fall short of our potential. If instead of describing dystopia, the majority of science-fiction was utopian—exploring the possible ways that things could go right for us—I believe it would, in a very real sense, point us a little bit more in that direction.
If that's a bit too lofty for you, another way to think about this is to imagine what your life could be like 100 years from now (i.e. if you'd been born 60 years from now). Depending on how things go, you could be scraping by with a group of other radiation-poisoned humans as the world recovers from nuclear holocaust. Or, you could be out exploring the galaxy in a luxury space yacht, with a potential lifespan in the centuries or millennia.
Which is more interesting to you? Which future would you rather live in?
This is the area I'm exploring with this series. I want to find the data and conduct the analyses that begins to show how our collective narrative (aliased by popular science-fiction) can bring about changes in our technological progress.
Of course this area is too large to explore in a single project, which is why I am doing it as a series. The first article in the series explored, at a very basic level, how technical terminology disperses through popular culture. You can find that article here: Tech Term Velocity.
In this project, print(fiction) the broad question I wanted to explore was this:
What separates fact from fiction?
...which is really just a cliché way of saying I wanted to explore the differences between nonfiction and fiction stories. My favorite method of consuming science-fiction is through books. Therefore, I chose to look at the differences between fiction and nonfiction books.
Without diving into the actual content of books, my goal was to look for patterns in the book metadata that could distinguish the two classes of books.
Imports and Configuration
# === General Imports === #
import pandas as pd
import numpy as np
from scipy.stats import randint, uniform
import matplotlib.pyplot as plt
import seaborn as sns
# === Configure === #
%matplotlib inline
pd.options.display.max_rows = 200
pd.options.display.max_columns = 200
# === ML Imports === #
# Preprocessing
import category_encoders as ce
from sklearn.preprocessing import StandardScaler
from sklearn.experimental import enable_iterative_imputer
from sklearn.impute import SimpleImputer, IterativeImputer
# Model validation
from sklearn.model_selection import train_test_split, cross_val_score, RandomizedSearchCV
from sklearn.pipeline import make_pipeline, Pipeline
from sklearn.metrics import confusion_matrix, accuracy_score, f1_score, classification_report
# Models
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
# Interpretations
import eli5
from eli5.sklearn import PermutationImportance
I searched around a bit for data that could help me answer this question. There were a few datasets on Kaggle that contained metadata scraped from GoodReads. I liked the idea of using metadata, but none of these datasets were quite what I wanted. Therefore, I decided to gather my own dataset and gain some more experience with web scraping along the way.
Scraping
I did not build the Scrapy scraper completely from scratch, though I probably would have if I hadn't run across a small project on GitHub that was built for this exact purpose. I figured the scraper itself wasn't as important to this particular project as the data it produced, so I decided to use it instead of building my own.
However, I did get my feet pretty wet with Scrapy in order to understand what it was doing and why it didn't work when I first tested it out. I forked it and made some minor modifications to the code to fix a bug or two and make it better fit my use-case. Overall, finding the project was a definite time-saver, and I still got to learn about how to build a robust web spider with Scrapy.
My fork of the repository can be found here: tobias-fyi/GoodReads.
Of course it's not realistic to simply send off the spider to crawl every single book on GoodReads. I decided to look at the popular reading lists that are curated on the site, as they do a good job aggregating books chosen by many tens of thousands of users.
The list I ended up choosing to use as the basis for my scrape is called Books That Everyone Should Read At Least Once, which has a little over 20,000 titles, voted on by almost 100,000 users.
This worked out nicely, and after about 16 hours of crawling and scraping, I had my dataset containing the metadata for about 20,000 books.
The raw output from the scraper took the form of a series of JSON files. I wrote a few helper functions that concatenated these files into a single Pandas DataFrame, with some basic initial preprocessing steps built in, such as dropping duplicate records and cleaning up column names.
The output of the scraper was a set of JSON files. In order to use it in the project, I'll need to convert to Pandas DataFrame.
In case you're curious, here are some functions I wrote to read and combine json files into single DataFrame and do some preprocessing.
def json_cat(json_files):
"""Reads and concatenates a list of .jl (json lines)
files into a single dataframe.
"""
# Read the books json files into a list of dataframes
dfs = [pd.read_json(filepath, lines=True) for filepath in json_files]
# Concatenate the list of dataframes
df = pd.concat(dfs, sort=False)
return df
def encode_book_genres(df):
"""Deconcatenates top 30 book genres into separate
features, OneHotEncoding style.
"""
# Set comprehension - creates a set of all genres listed in dataset
all_genres = {genre for row_genres in df["genres"] for genre in row_genres}
# Create a new feature for every genre
for genre in all_genres:
has_genre = lambda g: genre in g
df[genre] = df.genres.apply(has_genre)
# Create list of top 30 most common genres
most_common_genres = df[list(all_genres)].sum().sort_values(ascending=False).head(30)
most_common_genres = most_common_genres.index.tolist()
# Drop all but the top 30 genres from the dataframe
unwanted_genres = list(all_genres - set(most_common_genres))
df = df.drop(columns=unwanted_genres)
# Drop the original "genres" feature
df = df.drop(columns=["genres"])
return df
def book_pub_date(df):
"""Deconcatenates book publish_date to three separate features
for year, month, and day. Drops the original publish_date feature.
"""
# === The Pandas method === #
# Convert the "publish_date" column to datetime
df["publish_date"] = pd.to_datetime(df["publish_date"], errors="coerce", infer_datetime_format=True)
# Break out "publish_date" into dt components
df["publish_year"] = df["publish_date"].dt.year
df["publish_month"] = df["publish_date"].dt.month
df["publish_day"] = df["publish_date"].dt.day
df = df.drop(columns=["publish_date"]) # Drop the OG publish_date
return df
def book_cat(paths_list, output_filename):
"""Reads and concatenates a list of book_*.jl (json lines) files."""
# === Concatenate the list of dataframes === #
df = json_cat(paths_list)
# === Initial wrangling === #
df = df.drop_duplicates(subset=["url"]) # Drop duplicate records
# Format column names with pyjanitor
df = (df.clean_names())
# Break ratings_histogram (array) into component features
df_hist = df["rating_histogram"].apply(pd.Series)
rating_cols = {} # Dict for mapping column names
for col in df_hist.columns.tolist():
rating_cols[col] = f"{col}_rating_count"
# Rename according to mapper created above
df_hist = df_hist.rename(columns=rating_cols)
# Concat new columns onto main dataframe
df = pd.concat([df, df_hist], axis=1, join="outer")
# Drop extra column
df = df.drop(columns=["rating_histogram"])
df.to_csv(output_filename, index=False)
print(f"Created dataframe and saved to current directory as '{output_filename}'")
return df
books = book_cat(bookpaths, "must_read_books-01.csv")
print(books.shape)
books.head()
books.isnull().sum()
books.info()
Feature management
Before digging deep into the data, there was some initial processing and pruning to be done to the features to make them more manageable later on.
Right off the bat I removed some features that likely wouldn't prove useful in the model:
urlasin0_rating_countcharactersplaces
url and asin are obviously not going to be useful, just extra noise. As can be seen in the info table above, 0_rating_count was completely null because GoodReads doesn't allow books to get zero stars.
I based my decision to remove characters and places on my domain knowledge and on the quality of their data. In my experience, nonfiction books are much less likely to be set in a particular location or have characters.
On one hand, this could be valuable information for the model to know: if the book has a list of characters, it is more likely to be fiction. On the other hand, that information could be too useful—i.e. it could leak information about the target.
Both have a high proportion of null values—over 50%, as can be seen in the table above—and I cannot be sure whether the null values represent the fact that users simply haven't taken the time to add that data to those books, or if those books really do not have any significant characters or places.
drop_cols = [
"url",
"asin",
"0_rating_count",
"characters",
"places",
]
books = books.drop(columns=drop_cols)
genres has to happen here in order to complete the data type conversion directly below this.
books = books.dropna(subset=["genres"])
print(books.shape)
books.head(2)
Data types
I did some initial feature engineering early on to make the features more manageable for me and usable for the model. More feature engineering will take place later on.
First, the publish_date feature needed to be split up into its component parts (year, month, day), because Scikit-learn models can't directly use the datetime format. By splitting them up into integers, their meaning can be preserved and the model can process them.
Second, I had to deal with nested data. Most notably, the genres column was organized as an array of genres for each book. Because Pandas doesn't parse this kind of data by default, the column imported as the object (text) datatype. The best way I found to deal with it, without delving into natural language processing, was to break out each genre into its own column, one-hot encoding style.
This step was very important, as I used the genres to engineer the fiction target.
genre null values are removed.
def book_pub_date(df: pd.DataFrame):
"""Deconcatenates book publish_date to three separate features
for year, month, and day. Drops the original publish_date feature.
"""
# Convert the "publish_date" column to datetime
df["publish_date"] = pd.to_datetime(df["publish_date"], errors="coerce", infer_datetime_format=True)
# Break out "publish_date" into dt components
df["publish_year"] = df["publish_date"].dt.year
df["publish_month"] = df["publish_date"].dt.month
df["publish_day"] = df["publish_date"].dt.day
df = df.drop(columns=["publish_date"]) # Drop the OG publish_date
return df
books = book_pub_date(books)
books.head(2)
Null values
A feature with a large proportion of null values is far less likely to be useful, as imputing (filling in) the missing data can add noise—or even artificial patterns—that would adversely affect the model. Therefore, a feature such as that can usually be considered extraneous and removed from the dataset. With a small enough proportion of null values, imputing or otherwise filling in the missing data is more likely to preserve the preexisting patterns in those features.
Depending on the distribution of null values throughout the dataset and the characteristics of the features, it might be better to remove rows that have a high proportion of nulls.
There is one specific feature for which I want to drop all rows that have a null: 'genres'. Because I am going to be using this to engineer my target, I don't want to risk biasing the model by imputing the missing values.
After removing those, I was left with 18,573 rows.
The above table doesn't do a great job describing the null value situation. Below is a visualization (thanks to missingno) showing the distribution and proportion of null values per feature (after removing rows with null genres).
books.isnull().sum()
# import missingno as msno
msno.matrix(books)
plt.title("Null values by feature")
# Settings for saving as image
# plt.tight_layout()
# plt.savefig("null_values_by_feature.png", dpi=80)
I used Scikit-learn's IterativeImputer to impute the missing values for many of the features with null values. Basically, it models each feature as a function of the other features, using that model to "predict" what the missing value would have been if it wasn't null.
There are a couple of features that had to be dealt with differently, original_publish_year and series, as their null values actually held information.
For original_publish_year, I assumed that a null value indicated that the book had not been previously published. And similarly for series, I assumed null indicated a standalone book. I transformed both of these features into binary: 0 indicating the value was null, 1 indicating it was not.
# === Encode the 'genres' feature === #
def encode_book_genres(df: pd.DataFrame):
"""Deconcatenates top 30 book genres into
separate features, OneHotEncoding style.
"""
from ast import literal_eval
# === Convert 'genres' to python list === #
df["genres"] = df["genres"].apply(literal_eval)
# Create a set of all distinct genres listed in dataset
all_genres = {genre for row_genres in df["genres"] for genre in row_genres}
# Create a new feature for every genre
for genre in all_genres:
has_genre = lambda g: genre in g
df[genre] = df.genres.apply(has_genre)
# Create list of top 30 most common genres
# NOTE: I ended up only using 'fiction', the top result
most_common_genres = df[list(all_genres)].sum().sort_values(ascending=False).head(1)
most_common_genres = most_common_genres.index.tolist()
# Drop all but the top 30 genres from the dataframe
unwanted_genres = list(all_genres - set(most_common_genres))
df = df.drop(columns=unwanted_genres)
# Drop the original "genres" feature
df = df.drop(columns=["genres"])
# Convert from Boolean to binary
df = df.replace(to_replace={True: 1, False:0})
# Format column names with pyjanitor
df = (df.clean_names())
return df
books = encode_book_genres(books)
books.head(2)
books["series"] = books["series"].notnull().replace(to_replace={True: 1, False:0})
books["series"].value_counts()
books["republish"] = books["original_publish_year"].notnull().replace(to_replace={True: 1, False:0})
books = books.drop(columns=["original_publish_year"])
books["republish"].value_counts()
books.dtypes
books.describe(exclude="number").T.sort_values(by="unique")
books[books["title"] == "When Breath Becomes Air"]
Looks like there are some duplicates. I'm going to use title, author, and publish_year as the subset this time.
books = books.drop_duplicates(subset=["title", "author", "publish_year"])
books.shape
It looks like there were around 200 duplicates that slipped through the cracks. Not this time!
Distributions and outliers
As can be expected with media such as books, there were some large outliers with regards to popularity. While the vast majority of books had relatively few ratings and even fewer reviews, where is a relatively small group of authors that struck gold, with them and their books becoming household names.
I found it somewhat surprising that my dataset reflected this reality so closely, as I thought a list of "must reads" would be biased toward more popular books. That bias is likely present—the dataset is large enough to include many books that never became household names—though not to the degree I initially thought.
# Caption: Distribution of the number of ratings.
plt.figure(figsize=(16, 8))
sns.distplot(books["num_reviews"])
plt.title("Distribution of 'num_reviews'");
# plt.savefig("num_reviews_distplot.png", dpi=160)
The distributions of the number of ratings and reviews are both heavily skewed to the right. The box in the boxplot below indicates the interquartile range, and the line that sits just barely to the right indicates the top of the third quartile. Basically, anything above that line can be considered an outlier.
Another way to look at the dispersion of the data is to consider the mean versus the median, along with the standard deviation. In the case of num_ratings, the relative size of the mean (46,958, with a standard deviation of 212,398!) compared to the median (4,135) indicates that the mean is significantly influenced by outliers.
# Caption: Boxplot showing distribution of the number of ratings.
plt.figure(figsize=(16, 8))
sns.boxplot(x=books["num_ratings"])
plt.title("Distribution of 'num_ratings'");
# plt.savefig("num_ratings_boxplot.png", dpi=160)
Curious what those books are?
# Caption: Highest rated books.
hiratings = books.nlargest(20, ["num_ratings"]).set_index("title")["num_ratings"]
plt.figure(figsize=(16, 8))
sns.barplot(hiratings, hiratings.index, palette="deep")
plt.title("Books with the most ratings");
# plt.tight_layout()
# plt.savefig("books_most_ratings.png", dpi=160)
Dealing with the outliers
Notice anything about the books with the most ratings—i.e. the most popular?
Every one the top 12 most popular books is fiction.
Based on that observation, I figured the model would find these particular outliers to be useful. However, given the huge range of these features, they would have to be scaled for use in any linear models. To this end, I included an instance of Scikit-learn's StandardScaler in my pipeline, which standardizes numeric features, transforming the data such that each feature has a mean of zero and standard deviation of 1. Basically, that just brings all of the numeric data into the same range.
Scaling the data is very important for linear models or neural networks—not so much for decision tree-based models. Therefore, I only used the StandardScaler as needed.
# Caption: Boxplot showing the distribution of number of pages.
plt.figure(figsize=(16, 8))
sns.boxplot(x=books["num_pages"])
plt.title("Distribution of 'num_pages'");
# plt.savefig("num_pages_boxplot.png", dpi=160)
Another feature that had stark outliers was the number of pages. There were several books with over 5,000 pages when the majority had less than 500. As with the outliers discussed above, I figured that the number of pages was relevant to the model.
Upon further inspection, however, I found that most of the outliers in this case were not actually single books, but entire book series. Therefore, I decided to remove the farthest outliers—those over 2,000 pages in length.
At this point, I was left with 18,344 rows in my dataset.
# This could be one of the sliders on the dashboard
cutoff = 2000
books_over_pages_cutoff = books[books["num_pages"] > cutoff]
print(books_over_pages_cutoff.shape)
books_over_pages_cutoff.head(2)
books2 = books.drop(books_over_pages_cutoff.index, axis=0)
print(books2.shape)
books2.head(2)
books2["num_pages"].describe()
Feature relationships
Another line of inquiry I wanted to explore before diving into the model was relationships between features and between the target and the features. The obvious first step was to create a matrix of scatterplots between what I believed to be the most important features and the target. As the target is binary, I thought it best to represent it as a third dimension in each of the scatterplots: color.
# Caption: Matrix of scatterplots showing relationships between certain features.
sns.pairplot(
books2,
hue="fiction",
vars=["num_reviews", "avg_rating", "num_pages"],
palette="deep",
plot_kws=dict(alpha=0.8, s=20),
height=4,
);
Although it's difficult to see much in the way of detail, this matrix is great for getting an overall idea of what's going on between the features in question.
One characteristic that immediately stuck out to me is the majority of outliers in num_ratings are fiction books. This helps to confirm my earlier hypothesis that these outliers will be valuable to the model.
Also, I noticed that avg_rating seems to be different between fiction and nonfiction books. The comparison of distributions (middle-middle) and the scatter plots comparing avg_rating to num_pages (middle-bottom and middle-right) seem to indicate that nonfiction books are rated slightly higher than fiction.
Seeing this difference in densities when grouped by the target made me want to explore other features in this manner. An interesting one I found was publish_year. As can be seen in plot below, it seems that the "golden years" of fiction were in the mid-2000s (at least according to this reading list), whereas the mid-2010s had more good nonfiction.
# Caption: Distribution of books by fictionality.
sns.pairplot(
books2[books2["publish_year"] > 1960],
hue="fiction",
vars=["publish_year"],
palette="deep",
plot_kws=dict(alpha=0.8, s=20),
height=8,
);
Although the exploration above is relatively basic, I can see that there seem to be some meaningful relationships in the data. Most important to my purposes is confirming that there are patterns that indicate some differentiation in the features when grouping them by the target.
With that in mind, I felt good enough to get into playing with some models.
Baseline Models
As I discussed above, the most complex model is not necessarily the best model. It's generally a good idea to start with a simple baseline model and progressively add complexity in proceeding iterations. The chosen evaluation metric will (ideally) indicate when the added complexity is beneficial, and when it is not.
train, test = train_test_split(books, stratify=books["fiction"], test_size=0.2, random_state=92)
train, val = train_test_split(train, stratify=train["fiction"], test_size=0.2, random_state=92)
train.shape, val.shape, test.shape
target = "fiction"
# Arrange y vector
y_train = train[target]
y_val = val[target]
y_test = test[target]
print(y_train.shape, y_val.shape, y_test.shape)
# Arrange X matrices
X_train = train.drop(columns=[target])
X_val = val.drop(columns=[target])
X_test = test.drop(columns=[target])
print(X_train.shape, X_val.shape, X_test.shape)
Majority class
The simplest possible model is to simply predict the majority class every time, regardless of input. Assuming an evenly distributed binary target, that model should be right about half the time, or have a 50% accuracy. In my case, the majority class baseline is just north of that, around ~.52, or ~52%.
Now I know that no matter what model I end up choosing and what features I end up using, it must have an accuracy of more than .52. If I can't beat that...
print(y_train.value_counts())
sns.distplot(y_train);
maj = y_train.mode()[0] # Mode is 1 (fiction)
# Simply predict 1 for every training example
y_pred_maj = [maj] * len(y_train)
# Baseline accuracy
accuracy_score(y_train, y_pred_maj)
Limited logistic regression
Taking it a step further, I find it useful to get a second baseline using an algorithm with a bit more complexity than that, but far less complexity than what is possible given the problem parameters. In this case I chose to train a limited logistic regression model using only a few of the features: num_reviews, avg_rating, and num_pages. This simple baseline model had an accuracy of ~.63 and an F1 score of ~.65.
I will go into some detail on what the F1 score is in the Model Validation section. For now, suffice to say that the discrepancy between it and the accuracy is due to the slight imbalance of the classes in the target.
base_features = [
"num_reviews",
"avg_rating",
"num_pages",
]
# Arrange X matrices
X1_train = train[base_features]
X1_val = val[base_features]
X1_test = test[base_features]
X1_train.shape, X1_val.shape, X1_test.shape
pipe1 = Pipeline([
("scaler", StandardScaler()),
("imputer", SimpleImputer(strategy="median")),
("logreg", LogisticRegression(random_state=92)),
])
# Train base pipeline
pipe1.fit(X1_train, y_train)
y_pred1 = pipe1.predict(X1_val)
# Compute accuracy
print("Baseline accuracy:", accuracy_score(y_val, y_pred1))
confusion_matrix(y_val, y_pred1)
Default random forest
I actually trained one more baseline model to set the bar higher and get a more accurate and precise idea of how high the bar will go given the problem, the available features, and the amount of data in the dataset.
This last baseline was a random forest that I left with default hyperparameters, trained on the full set of features as they were at this point in the process. This more complex—maybe even overly complex—model did quite a bit better, with an accuracy of ~.73 and an F1 of ~.75.
That's the score to beat as I iterate on the model and add complexity.
def_drop_columns = [
"title",
"author",
"language",
]
X2_train = X_train.drop(columns=def_drop_columns)
X2_val = X_val.drop(columns=def_drop_columns)
X2_test = X_test.drop(columns=def_drop_columns)
rf1_pipe = Pipeline([
("encoder", ce.OrdinalEncoder()),
("imputer", SimpleImputer(strategy="median")),
("rfc", RandomForestClassifier(random_state=92)),
])
# Train default random forest
rf1_pipe.fit(X2_train, y_train)
# Made predictions to get validation accuracy
y_pred_rf1 = rf1_pipe.predict(X2_val)
# Compute evaluation metrics
print("Default random forest eval metrics:")
print(" Accuracy:", accuracy_score(y_val, y_pred_rf1))
print(" F1 score:", f1_score(y_val, y_pred_rf1))
Model validation
As always, before jumping into building and training I needed to make some decisions about how I will measure success—or, more accurately, how the model measures success. Choosing appropriate evaluation metrics is crucial to get an accurate understanding of how well a model is fitting (or overfitting) the data. An overly complex model will likely overfit the training data, leading to a model that performs poorly on new inputs on which is was not trained. Decision trees are particularly prone to overfitting
Target distribution
The target I'm building a model to predict is binary, 1 or 0; fiction or nonfiction. Therefore, the metrics I choose must be appropriate for a binary classification model. The most common way to measure the performance of a binary classifier is the accuracy score—i.e. the proportion of predictions correctly classified by the model.
A very important characteristic to look at when choosing a metric is the distribution of the target variable. In the case of binary classification, if the target is skewed one way or the other, accuracy is not a good method of evaluating performance.
Although the target in my dataset is very evenly distributed between fiction (~54%) and nonfiction (~46%), it still is skewed a little bit. This means that if I use accuracy, I can expect it to be a decent indicator of performance, but slightly less than ideal. The reason for this is that accuracy only tracks the number of the model's mistakes, irrespective of how the model made those mistakes.
Types of errors
In the case of binary classification, there are two ways a prediction can be wrong: false positive (type I error) or false negative (type II error). As the name suggests, false positive is when the model (falsely) predicts positive or 1 when the actual label is 0. False negative is the alternative to that, when the model predicts negative or 0 when the label is 1.
A better method of evaluating performance will take into account the number of each type of error in a model's predictions. Depending on the target and what the model will be used for, it may be better to use a metric that looks at one or the other.
A common example is a spam detection model. In this case, it is generally better to let some spam emails through the filter into the users' inboxes (false negative) than to put potentially important emails into the spam folder (false positive).
Confusion matrix
A common and effective way to get deeper insight into a classification model's predictions is with a confusion matrix, which is a type of contingency table with the target's classes laid out along both axes, the rows representing the actual classes and the columns showing the predicted classes.
# Caption: Confusion matrix for default random forest model.
from sklearn.utils.multiclass import unique_labels
unique_labels(y_val) # Create unique labels
def plot_confusion_matrix(y_true, y_pred):
labels = unique_labels(y_true)
columns = [f'Predicted {label}' for label in labels]
index = [f'Actual {label}' for label in labels]
table = pd.DataFrame(confusion_matrix(y_true, y_pred),
columns=columns, index=index)
return sns.heatmap(table, annot=True, fmt='d', cmap='viridis')
# Plot the confusion matrix
plt.figure(figsize=(10, 8))
plt.title("Confusion matrix: default random forest")
font = {'family' : 'normal',
'weight' : 'bold',
'size' : 14}
plt.rc('font', **font)
plot_confusion_matrix(y_val, y_pred_rf1);
Where each row and column intersect, it shows the number of predictions that fall into that category. In the case of binary classification, this results in a table containing four numbers: true positives, true negatives, false positives, and false negatives. A classifier that made no mistakes would have non-zero numbers only on the main diagonal (top left to bottom right).
Evaluation metrics
There are three main metrics that can be derived from the confusion matrix. The first is precision, which is the proportion of the total positives (true positive + false positive) that were correctly predicted to be positive (true positive). The second is recall, which is the proportion of the total predicted values (true positives + false negatives) that were correctly predicted to be positive (true positive). The last metric I'll be covering now is the F1 score: the weighted average of precision and recall.
I chose to use the F1 score as the primary evaluation metric for my model because I'm more interested in reducing the number of mistakes in general, as opposed to preferring to optimize for one or the other.
true_pos = 1156
false_pos = 400
precision = true_pos / (true_pos + false_pos)
print(f"Precision: {precision}")
true_pos = 1156
false_neg = 380
precision = true_pos / (true_pos + false_neg)
print(f"Precision: {precision}")
from sklearn.metrics import classification_report
print(classification_report(y_val, y_pred_rf1))
Let's see what we can do to increase that score.
Feature Engineering
When I initially started this project, I went through the process of validating and training a model or two that tried to predict the average rating of books. This was by far the most common target chosen by those who started Kaggle kernels using other GoodReads datasets. Although this may have the most obvious business value if I was a data scientist working for a book publisher, to me this wasn't a particularly interesting target to try to predict.
Target practice
I realized this when I hit a wall with my progress in improving the rating-predictor model. One reason was that I did not see any obvious useful features that could be engineered. However, once I found my way to the idea of predicting the fictionality of the books, the target drove the direction I took with my feature engineering. It was a great learning experience for me in engineering features toward the specific target that the model is trying to predict. I called this process "target practice".
Here are the feature ideas I came up with and engineered (all in short succession once the new target was chosen):
- Title begins with "The"
- Has subtitle: contains ":"
- Title character count
- Title word count
- Title longest word
- Author number of names
- Author middle initial
- Ratings (stars) ratio: 1 + 2 / 4 + 5
def engineer_features(data):
"""Engineer a handful of new features."""
# Create new feature that is if the title begins with "The"
data["the_title"] = data["title"].str.startswith("The")
# New feature - has_subtitle
data["has_subtitle"] = data["title"].str.contains(":")
# New feature - title character length
data["title_char_count"] = data["title"].apply(lambda x: len(x))
# New feature - title word count
data["title_word_count"] = data["title"].apply(lambda x: len(x.split()))
# New feature - title longest word
data["title_longest_word"] = data["title"].apply(lambda x: len(max(x.split(), key=len)))
# New feature - author number of names
data["author_name_count"] = data["author"].apply(lambda x: len(x.split()))
# New feature - author middle initial
pat = r"\w* (\w. )+ \w*"
data["author_middle_initial"] = data["author"].str.contains(pat, regex=True)
# New feature - low/high rating ratio
data["rating_ratio"] = (data["1_rating_count"] + data["2_rating_count"]) / (data["4_rating_count"] + data["5_rating_count"])
# Replace Boolean with binary
data = data.replace(to_replace={True: 1, False:0})
return data
Forest with new features
To get an initial idea of the effect that these new features had on the model, I retrained a new random forest with all the same hyperparameters (defaults). The result was a significant boost in the model's F1 score, from ~.75 to ~.79. That made me happy.
However, because I engineered all of the new features at once—i.e. I didn't retrain the model after every one—this does not give me insight into which ones were useful. In fact, at this point I hadn't looked at how useful any of the specific features were.
X3_train = engineer_features(X_train)
X3_val = engineer_features(X_val)
X3_test = engineer_features(X_test)
rf2_pipe = Pipeline([
("encoder", ce.OrdinalEncoder()),
("imputer", SimpleImputer(strategy="median")),
("rfc", RandomForestClassifier(random_state=92)),
])
# Train default random forest
rf2_pipe.fit(X3_train, y_train)
# Made predictions to get validation accuracy
y_pred_rf2 = rf2_pipe.predict(X3_val)
# Compute evaluation metrics
print("Default random forest eval metrics:")
print(" Accuracy:", accuracy_score(y_val, y_pred_rf2))
print(" F1 score:", f1_score(y_val, y_pred_rf2))
Permutation Importances
It is likely that some of the features do not help the model make correct predictions. Indeed, some may even be worse than that: they could add noise that makes the model perform worse.
To address this potential problem, I'm going to find each feature's importance to the model using a method called permutation importance. Basically, this method will go through each of the features, one by one, replacing their data with random noise generated from the distribution of the original data. The performance of the model will be evaluated and compared with the score using all of the original data to find the effect that each feature has on the performance of the model.
The following table is the result of running permutation importance on both the new and old features.
# Use the same (fitted) steps from main pipeline
transformers = Pipeline([
("encoder", rf2_pipe.named_steps["encoder"]),
("imputer", rf2_pipe.named_steps["imputer"]),
])
# Encode and impute
X3_train_transformed = transformers.transform(X3_train)
X3_val_transformed = transformers.transform(X3_val)
permuter = PermutationImportance(
rf2_pipe.named_steps["rfc"],
scoring='f1',
n_iter=5,
random_state=42
)
permuter.fit(X3_val_transformed, y_val)
feature_names = X3_val.columns.tolist()
pd.Series(permuter.feature_importances_, feature_names).sort_values(ascending=False)
eli5.show_weights(
permuter,
top=None, # Show permutation importances for all features
feature_names=feature_names
)
Along with being useful for feature selection, I find it interesting to see what features have the largest positive effect on the model's predictive power. From this table, I can see that the majority of the benefit I got from engineering the new features came from has_subtitle. This feature, according to the permutation importance table, is the most important predictor of the lot, and simply indicates whether the title of the book has a colon in it. My intuition was that having a subtitle is very common for nonfiction books, not so much for fiction. It seems that my intuition was generally good.
more_drop_cols = [
"publish_month",
"author",
"title_word_count",
"title",
"publish_day",
]
# New features are already engineered
X4_train = X3_train.drop(columns=more_drop_cols)
X4_val = X3_val.drop(columns=more_drop_cols)
X4_test = X3_test.drop(columns=more_drop_cols)
rf3_pipe = Pipeline([
("encoder", ce.OrdinalEncoder()),
("imputer", SimpleImputer(strategy="median")),
("rfc", RandomForestClassifier(random_state=92)),
])
# Train default random forest
rf3_pipe.fit(X4_train, y_train)
# Made predictions to get validation accuracy
y_pred_rf3 = rf3_pipe.predict(X4_val)
# Compute evaluation metrics
print("Default random forest:")
print(" Accuracy:", accuracy_score(y_val, y_pred_rf3))
print(" F1 score:", f1_score(y_val, y_pred_rf3))
Once I removed those features, I saw another bump of ~.01 in the model's F1 score. That put the model up to an F1 score just south of .80 on the validation set.
Now that I'd engineered some new features and pruned them to only the useful ones, it was time to iterate and tune.
Cross-validation
Up until this point, I had been using a consistent train-test-validation set split. However, now that I was going to be tuning hyperparameters, it made sense to start using cross-validation. I won't get into the details of what that is in this particular article. If you're curious, search for it on Duck Duck Go, or read about it in The Python Data Science Handbook.
# Start from original dataset, because data
# will only be split into train and test
books2 = engineer_features(books)
print(books2.shape)
books2.head(2)
train, test = train_test_split(books2, stratify=books2["fiction"], test_size=0.2, random_state=92)
train.shape, test.shape
# No val this time bc using cross-validation
target = "fiction"
drop_cols = [ # Columns not useful to model
"title",
"author",
"language",
"publish_month",
"publish_day",
]
# Arrange y vector
y_train = train[target]
y_test = test[target]
print(y_train.shape, y_test.shape)
# Arrange X matrices
X_train = train.drop(columns=[target] + drop_cols)
X_test = test.drop(columns=[target] + drop_cols)
print(X_train.shape, X_test.shape)
Hyperparameter tuning
Hyperparameters are the parameters of the model that are not directly learned during the training process—they must be adjusted manually. My preferred method of tuning them is with a randomized search.
Essentially, I provide a list of algorithm hyperparameters and a search window or distribution for each one. The algorithm runs through a specified number of iterations, each time randomly selecting values for each hyperparameter from their respective distribution and using those to train a new model. Each model is evaluated after each iteration using cross-validation. Then, once the search is over, the pipeline is refitted using the values that resulted in the model with the best cross-validation score.
As the search is random and limited to a reasonable number of iterations, some strategy is involved to find optimal search windows/distributions for each hyperparameter. The method I used for this is to start out searching a wide range of values and go through the process a few times, progressively narrowing the range around the values that come up as consistently optimal.
Validation
Before getting too deep into tuning the hyperparameters of a specific algorithm (it is computationally expensive to run exhaustive searches), I thought it best to test out a few different algorithms. I started out with the assumption that random forest or gradient-boosting would provide the best results, but it's generally good to test assumptions.
Though I did not test a very wide range of algorithms, I figured it would be worth it anyways to see if any of the more commonly-used ones showed any promise.
Somewhat to my surprise, the best models from tuning and cross-validation were basically on par with the default, non-cross-validated random forest.
The fact that the baseline was trained without using cross-validation could be one reason. In other words, the best model from the search might generalize better to new data; it could outperform the default one on the test dataset.
Or, it could be a result of the parameters and their ranges I searched—i.e. not giving a wide enough initial distribution. However, the wider the distribution, if the number of iterations is relatively low, the more "noise" might be present in the results of the search. I wanted to wait until I chose an algorithm to try increasing the number of search iterations significantly.
Here is the full list of algorithms I trained during the validation and iteration process, and their best F1 score:
- Logistic regression: ~.76
- K-nearest neighbors: ~.69
- Random forest: ~.80
- Gradient-boosted decision tree: ~.80
# Tune hyperparameters using cross-validation
rf3_pipe = Pipeline([
("encoder", ce.OrdinalEncoder()),
("imputer", IterativeImputer(random_state=92)),
("rfc", RandomForestClassifier(random_state=92)),
])
rf3_params = {
"imputer__initial_strategy": ["median", "most_frequent"],
"imputer__max_iter": randint(16, 40),
"imputer__tol": uniform(0.001, 0.05),
"imputer__n_nearest_features": randint(2, 10),
"imputer__imputation_order": ["ascending", "roman", "random"],
"rfc__n_estimators": randint(80, 300),
"rfc__max_depth": randint(6, 32),
"rfc__min_samples_split": uniform(0, 1),
}
# Define the search using parameter distros above
rf3_search = RandomizedSearchCV(
rf3_pipe,
param_distributions=rf3_params,
n_iter=5,
cv=5,
scoring='f1',
verbose=10,
return_train_score=True,
n_jobs=-1,
random_state=92,
)
# Train default random forest
rf3_search.fit(X_train, y_train)
# Best combination of hyperparameters and their resulting f1 score
print("Best hyperparameters", rf3_search.best_params_)
print("F1 score", rf3_search.best_score_)
nn_pipe = Pipeline([
("encoder", ce.OrdinalEncoder()),
("imputer", IterativeImputer(random_state=92)),
("nn", KNeighborsClassifier()),
])
nn_params = {
"imputer__initial_strategy": ["median", "most_frequent"],
"imputer__max_iter": randint(16, 40),
"imputer__tol": uniform(0.001, 0.05),
"imputer__n_nearest_features": randint(2, 10),
"imputer__imputation_order": ["ascending", "roman", "random"],
"nn__n_neighbors": randint(2, 20),
"nn__weights": ["uniform", "distance"],
"nn__algorithm": ["ball_tree", "kd_tree"],
"nn__leaf_size": randint(20, 50),
}
# Define the search using parameter distros above
nn_search = RandomizedSearchCV(
nn_pipe,
param_distributions=nn_params,
n_iter=5,
cv=5,
scoring="f1",
verbose=10,
return_train_score=True,
n_jobs=-1,
random_state=92,
)
# Train default random forest
nn_search.fit(X_train, y_train)
# Best combination of hyperparameters and their resulting f1 score
print("Best hyperparameters", nn_search.best_params_)
print("F1 score", nn_search.best_score_)
It seems that Random Forest is quite a bit better of an algorithm for this problem than k-nearest neighbors. Therefore, I won't be moving forward with nearest neighbors.
The last algorithm I'll try is a gradient-boosted decision tree classifier from XGBoost: XGBClassifier.
Gradient Boosting
Training a gradient-boosted decision tree using XGBoost.
Though I don't have a record of every single iteration of the below classifier search, the method I used to tune is to basically look at the values of each parameter, and moved the search range to more closely fit around those values.
I was surprised to find that my initial attempts at training the XGBClassifier had about the same performance as the default random forest with the newly-engineered features.
As I mentioned above, one hypothesis of what was causing the discrepancy (or lack thereof: I assumed gradient-boosting would increase the performance, which maybe wasn't a sound assumption), could be the simple fact that the randomized search doesn't cover every possibility. To test this, I increased the number of iterations and let 'er rip!
from xgboost import XGBClassifier
xgb1_pipe = Pipeline([
("encoder", ce.OrdinalEncoder()),
("imputer", IterativeImputer(random_state=92)),
("xgb", XGBClassifier(random_state=92)),
])
xgb1_params = {
"imputer__initial_strategy": ["median", "most_frequent"],
"imputer__max_iter": randint(16, 45),
"imputer__tol": uniform(0.02, 0.04),
"imputer__n_nearest_features": randint(2, 10),
"imputer__imputation_order": ["ascending", "roman", "random"],
"xgb__n_estimators": randint(80, 160),
"xgb__max_depth": randint(18, 48),
"xgb__learning_rate": uniform(0.05, .5),
}
# Define the search using parameter distros above
xgb1_search = RandomizedSearchCV(
xgb1_pipe,
param_distributions=xgb1_params,
n_iter=10,
cv=4,
scoring="f1",
verbose=10,
return_train_score=True,
n_jobs=-1,
random_state=92,
)
# Train default random forest
xgb1_search.fit(X_train, y_train)
# Best combination of hyperparameters and their resulting f1 score
print("F1 score", xgb1_search.best_score_)
print("Best hyperparameters:")
for param, val in xgb1_search.best_params_.items():
print( f"{param}: {val}")
Final algorithm
Even with 40 total fits (4 cross-validation folds, 10 iterations) the gradient-boosted classifier did not really outperform the random forest by any significant margin. Given the additional complexity and computation required for an XGBoost (gradient-boosting) model, I decided to move forward with the random forest classifier.
To continue testing the hypothesis that my initial number of iterations was too low for the search to converge on a good combination of hyperparameters, I trained more random forests with higher numbers of search iterations.
To my continued surprise, even after many, many more iterations, the absolute best F1 score I got still hovered right around ~.80.
rf4_pipe = Pipeline([
("encoder", ce.OrdinalEncoder()),
("imputer", IterativeImputer(random_state=92, n_nearest_features=3)),
("rfc", RandomForestClassifier(random_state=92)),
])
rf4_params = {
"imputer__initial_strategy": ["median", "most_frequent"],
"imputer__max_iter": randint(8, 20),
"imputer__tol": uniform(0.01, 0.04),
"imputer__imputation_order": ["ascending", "roman", "random"],
"rfc__n_estimators": randint(140, 200),
"rfc__max_depth": randint(6, 18),
"rfc__min_samples_split": randint(6, 14),
"rfc__min_impurity_decrease": uniform(0, .01),
}
# Define the search using parameter distros above
rf4_search = RandomizedSearchCV(
rf4_pipe,
param_distributions=rf4_params,
n_iter=15,
cv=5,
scoring='f1',
verbose=10,
return_train_score=True,
n_jobs=-1,
random_state=92,
)
# Train default random forest
rf4_search.fit(X_train, y_train)
# Best combination of hyperparameters and their resulting f1 score
print('F1 score', rf4_search.best_score_)
print('Best hyperparameters:')
for param, val in rf4_search.best_params_.items():
print( f"{param}: {val}")
rf4 = rf4_search.best_estimator_
y_pred_test_rf4 = rf4.predict(X_test)
# Compute evaluation metrics
print("Random forest 4 eval metrics:")
print(" Accuracy:", accuracy_score(y_test, y_pred_test_rf4))
print(" F1 score:", f1_score(y_test, y_pred_test_rf4))
y_pred_test_rf4 = rf4_search.predict(X_test)
# Compute evaluation metrics
print("Random forest 4 eval metrics:")
print(" Accuracy:", accuracy_score(y_test, y_pred_test_rf4))
print(" F1 score:", f1_score(y_test, y_pred_test_rf4))
plt.figure(figsize=(14, 12))
plt.title("Confusion matrix: final random forest, test data")
font = {'family' : 'normal',
'weight' : 'bold',
'size' : 22}
plt.rc('font', **font)
plot_confusion_matrix(y_test, y_pred_test_rf4);
train_id = X_train.reset_index()["index"]
test_id = X_test.reset_index()["index"]
test_id.head()
# Process the test data
transformers_2 = Pipeline([
("encoder", rf4_search.best_estimator_["encoder"]),
("imputer", rf4_search.best_estimator_["imputer"]),
])
# Encode and impute
X_test_transform = transformers_2.transform(X_test)
class_index = 1
# Make predictions with the trained random forest
y_pred_proba_rf4 = rf4_search.predict_proba(X_test_transform)[:, class_index]
# ROC AUC score ranges from 0-1; higher is better
print(f'Test ROC AUC for class {class_index}:')
print(roc_auc_score(y_test, y_pred_proba_rf4))
X_test = X_test.reset_index()
X_test.head()
X_test.shape, test_id.shape, y_pred_test_rf4.shape, y_pred_proba_rf4.shape, y_test.shape
y_test.reset_index().head()
# Create new dataframe to compare the predictions to the actual
df = pd.DataFrame({
"index": test_id,
"pred": y_pred_test_rf4,
"pred_proba": y_pred_proba_rf4,
})
print(df.shape)
df.head()
df = df.merge(y_test.reset_index())
print(df.shape)
df.head()
df = df.merge(
X_test,
how='left'
)
print(df.shape)
df.head()
df_wrong = df[df["pred"] != df["fiction"]]
print(df_wrong.shape)
df_wrong.head()
df_wrong = df_wrong.merge(books.iloc[df_wrong["index"]]["title"].reset_index())
print(df_wrong.shape)
df_wrong.head()
df_wrong.sample(n=60, random_state=92).sort_values(by="pred_proba")
Predicted probabilities
One way to interpret a trained model is to inspect the predictions that the model made on the test set, either individually or through various descriptive statistics and visualizations. A key component to this is the probabilities underlying each of the predictions.
By looking at predicted probabilities, I can look at instances when the model was very sure or not sure of its predictions, and if those predictions were correct or not. By looking at the cases where the model made mistakes, I can hopefully pick out patterns and glean some insight why the model made those mistakes.
If I find that the model consistently makes mistakes that don't make much sense, that could mean there is some more optimization to be done or weakness to be addressed.
False negatives
The first one that jumped out at me was subtitles. The model finds that feature very useful, yet of course there are some fiction books with subtitles. This mistake makes sense to me, and for the benefit that the feature adds to the model, it is worth incorrectly classifying some fiction books as nonfiction. One way to get around this would be to engineer another feature or three that attempts to be the complement to that feature, catching the instances when fiction books have subtitles.
Another feature that causes a similar type of error is the title character count. According to the feature importances table (and plot generated from the table's data, shown below) of the final random forest model, 'title_char_count' is also a very important feature. I can see that many of the false negatives (predicted nonfiction, actually fiction) have a high title character count.
rf4 = rf4_search.best_estimator_["rfc"]
importances = pd.Series(rf4.feature_importances_, X_train.columns)
# Plot feature importances
n = 20
plt.figure(figsize=(10,n/2))
plt.title(f'Top {n} features')
importances.sort_values()[-n:].plot.barh(color='grey');
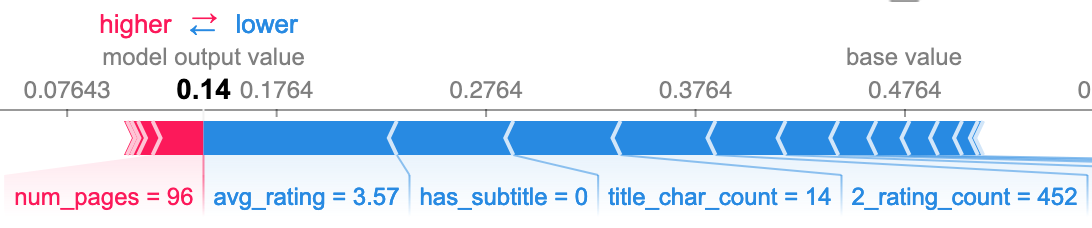
The following SHAP plots formalize my observations and provide more insight by showing the effect that the value of each feature had on the model's prediction. The following group are all false negative predictions, meaning the model predicted nonfiction when it was actually fiction.
I also wanted to see what features caused false negatives when the book had neither a high character count nor a subtitle. The third plot below shows the top three as republish, avg_rating, and publish_year. I was surprised to see republish, as it does not seem to be all that important to the model overall. The other two seem to be relatively good predictors, particularly avg_rating.
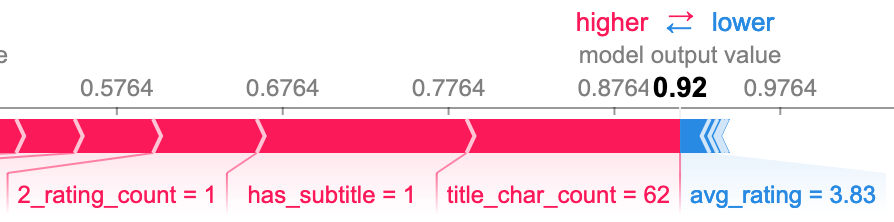
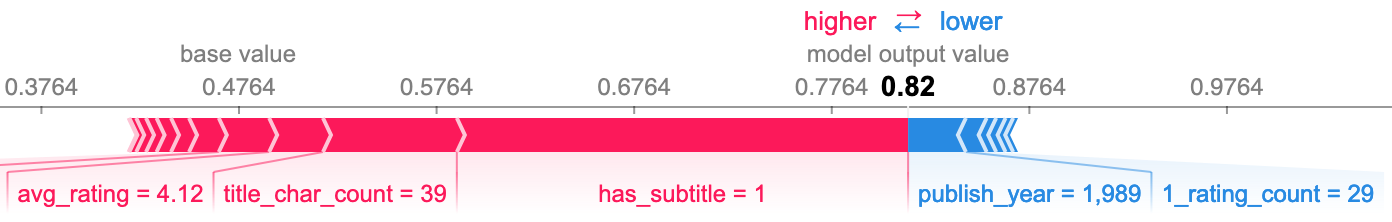
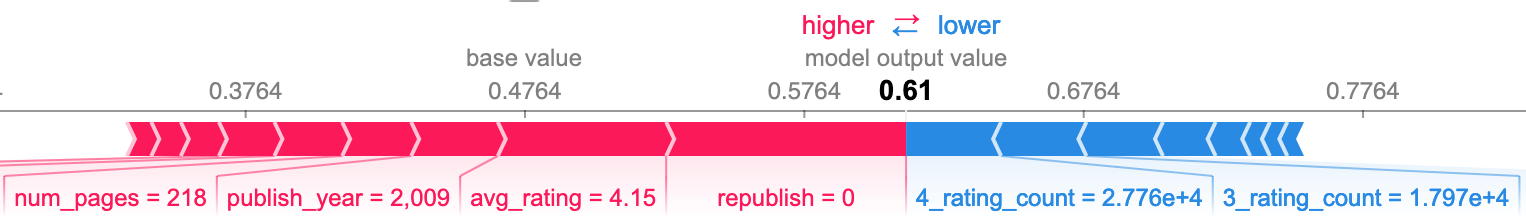
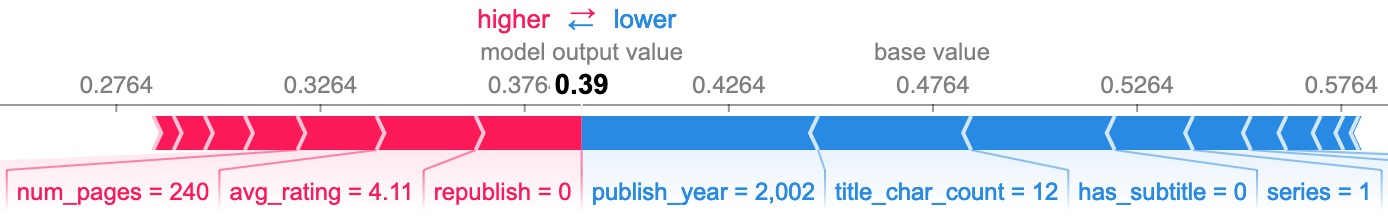
False positives
The other side of the confusion matrix again corroborates my initial observation. Many of the same features that provided some additional predictive power also mean some additional mistakes. However, the overall effect was net positive. If it wasn't these features causing the incorrect predictions, it would be others, and there would be more incorrect predictions overall.
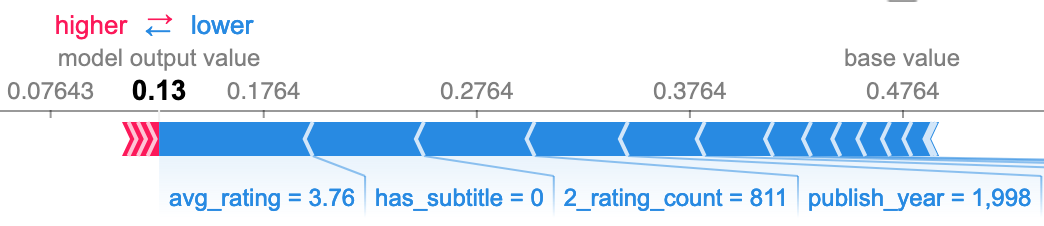
Deployment
So there you have it, the process of training a machine learning model, from start to finish. The only thing left to do is to deploy it.
I thought it would be interesting to be able to set up a dashboard that can be used to give dynamic inputs to the model and see the resulting prediction. I decided to build the dashboard using Plotly Dash and deploy it to Heroku.
So you don't have to scroll all the way up, here's another link to the live app: print(fiction).
Shoutout to HTML5UP for the template on which I based the design of the app. I made some minor modifications to it, but most of the design was straight from one of his templates.
If you made it this far, I salute you.
As always, thank you for reading and I'll see you in the next one!